Fantastic Info About How To Detect Snp
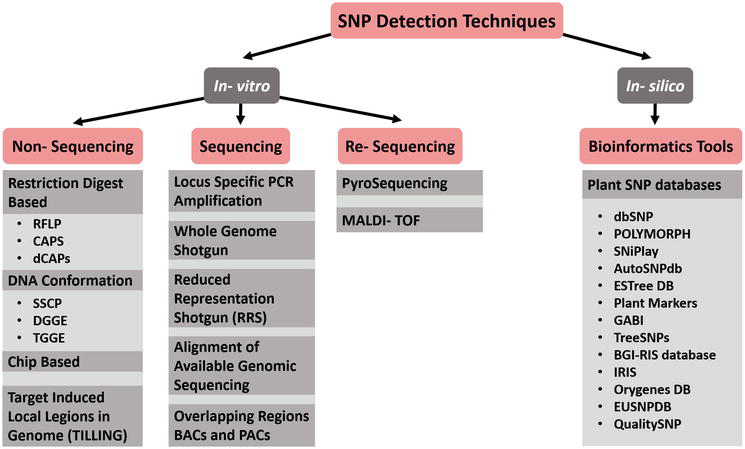
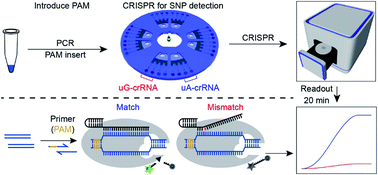
There are several methods to detect snps:
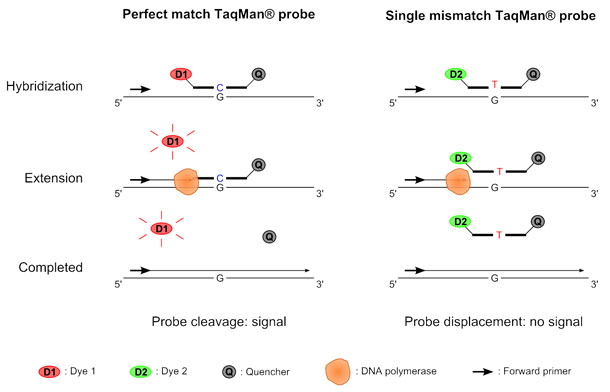
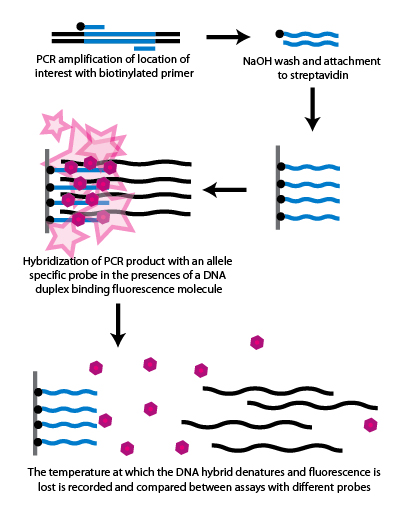
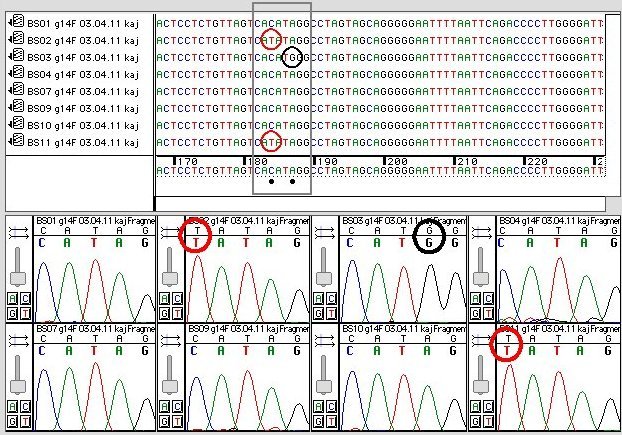
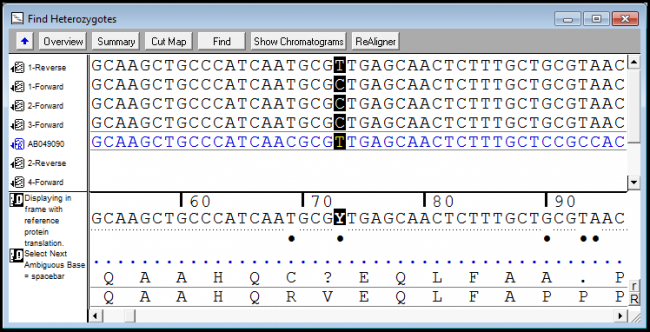
How to detect snp. You can use sequencher for comparative sequence alignments among a. This allows us to differentiate homozygous from heterozygous samples [5]. Sequencher has several powerful tools to help you detect mutations and snps in your dna sequences.
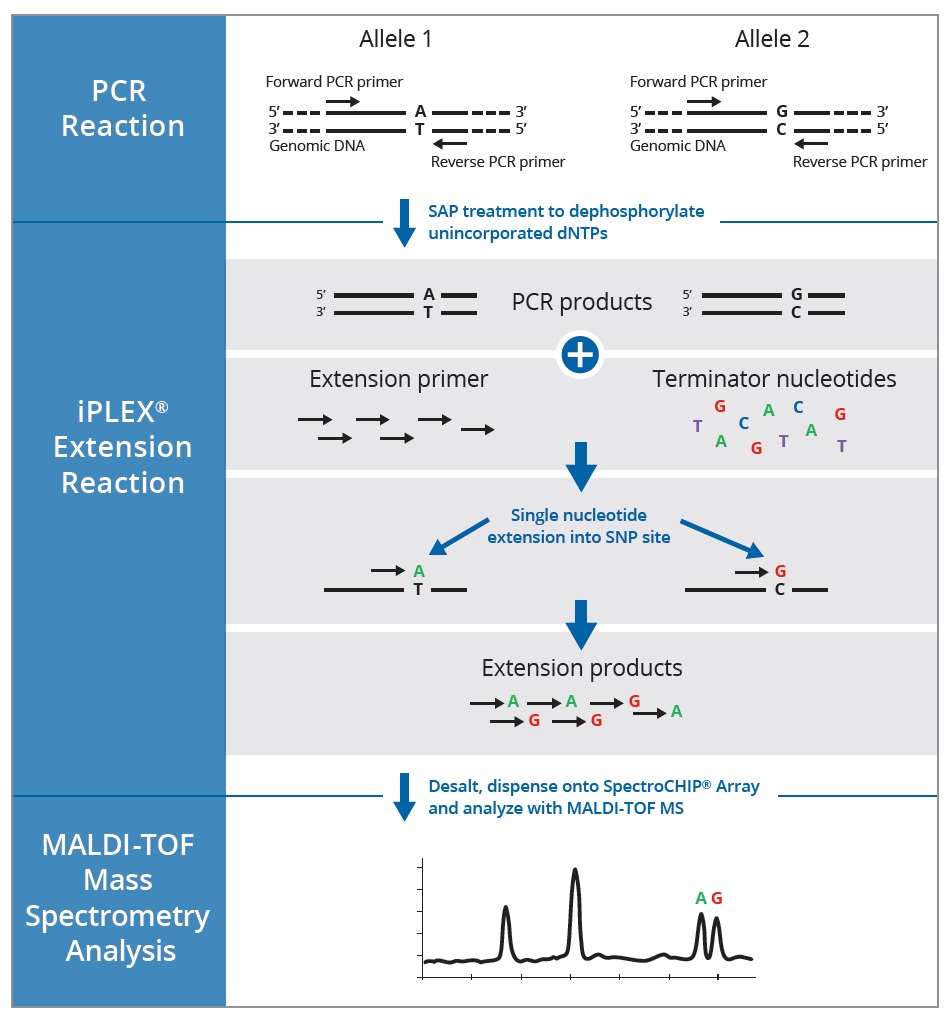
What is the best way to detect snps? Their characteristics, how snp markers are developed? Wicell currently utilizes the cytosnp 850k microarray from illumina (illumina™ infinium snp microarrays).
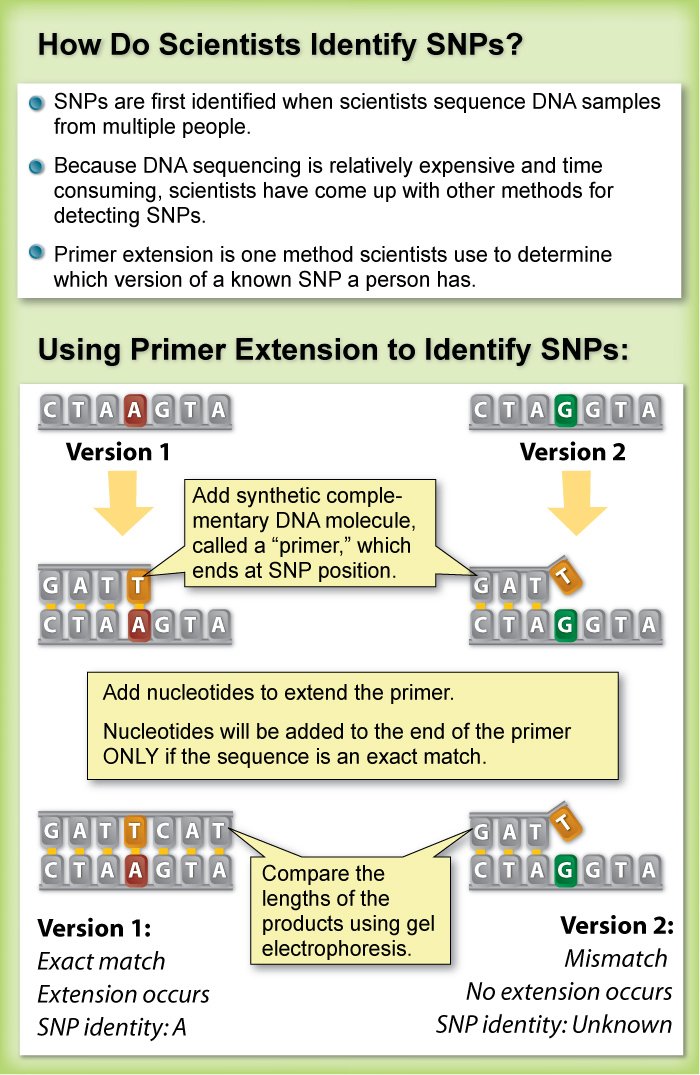
Restriction fragment length polymorphism (rflp) is considered to be the simplest and earliest method to detect snps. Next you will need to go to a website called ensembl. Follow general practices for the design of pcr primers.
What are snp markers, why they are so popular? Snp microarray uses known nucleotide sequences as probes to hybridize with the tested dna sequences, allowing qualitative and quantitative snp analysis through signal. Find your snps on the chromosome.
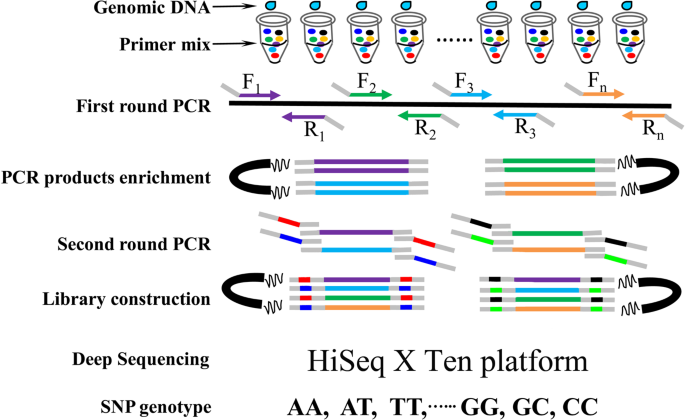
Methods of snp detection will be covered in this video.*. We have designed a snp identification pipeline. Any information about the time required and about the costs of each one is also.
In testing the performance of mrsnp on snps experimentally validated to affect mirna binding, mrsnp correctly identified 69% (11/16) of the snps disrupting binding. Samples are then analyzed using illumina bluefuse software and compared to a. Thus, a fast, economical and reliable sg embedded sensor is introduced to detect snp.